Performance Tools
Comparison of Call Graph Generation by Profiling Tools
Analyzing and optimizing the performance of parallel programs is critical to obtaining high efficiency on high performance computing (HPC) architectures. The complexity in hardware architectures and system software makes measuring and recording performance data challenging. Even so, a plethora of profiling tools exists for gathering and analyzing performance data. These profiling tools generate call graphs, which provide critical information about the program structure to analyze the performance of a program.
In [1], we compare several popular tools that are used in the HPC community – Caliper, HPCToolkit, Score-P, and TAU in terms of their runtime overheads, memory usage, and the size, correctness and quality of the generated call graph data. We conduct experiments on a parallel cluster by profiling three different proxy applications, AMG, LULESH, and Quicksilver, using both instrumentation and sampling under different sampling intervals and different numbers of processes. In order to analyze callpaths, we used and improved Hatchet, which is a performance analysis tool we develop in our group.
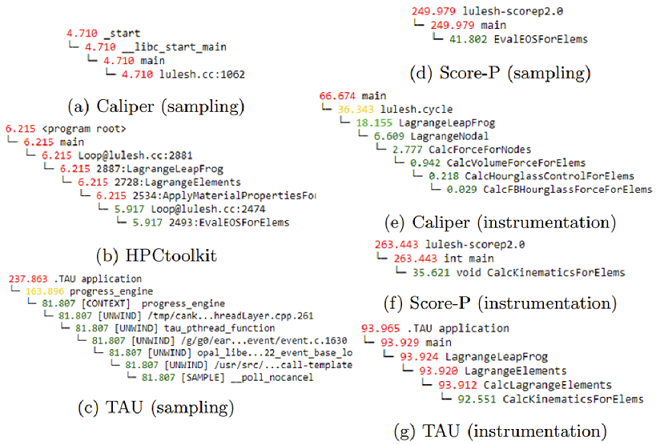
The above figure shows the callpath of the second slowest node generated by each tool using sampling and/or instrumentation. As can be seen, each tool usually identifies a different node as the second slowest, and in some cases, although the identified second slowest node is the same, its call path is different for different tools (e.g. Score-P instrumentation and TAU instrumentation).
For more details such as the methodology and evaluation for comparing runtime overhead, memory usage, and the size, correctness, and quality of the generated call graph data please see this link. please see “Comparative Evaluation of Call Graph Generation by Profiling Tools”.
Related Publications
[1] Onur Cankur et al, "Comparative Evaluation of Call Graph Generation by Profiling Tools", International Conference on High Performance Computing. Springer, May 2022